The interface is generated by some software called Zoom Search, and the interface is here. As an example, here is a query for the spider Enoplognatha latimana.
But...
Having an easy way to search a site using a URL API such as Open Search is great, but the feed is RSS 2.0, and as a result has very little information. For example, here's an extract:
<item>
<title>The Nearctic Spider Database: Enoplognatha latimana Hippa & Oksala, 1982 Description</title>
<link>http://canadianarachnology.dyndns.org/data/spiders/7561</link>
<description>THERIDIIDAE: Enoplognatha latimana taxonomic and natural history description in the Nearctic Spider Database.</description>
<zoom:context> ... Descriptions Home Search: Register Log in Enoplognatha latimana Hippa& Oksala, 1982 Temporary ... 2007 Arachnid Calendar FAMILY: THERIDIIDAE Sundevall, 1833 Genus: Enoplognatha Pavesi, 1880 ...</zoom:context>
<zoom:termsMatched>2</zoom:termsMatched>
<zoom:score>1804</zoom:score>
</item>
This information is intended to be displayed in a feed reader, and hence viewed by a human. But, what if I want to put this information in a database, or combine it with other data sources in a mashup, such as iSpecies? Well, I have to scrape information out of free formatted text. In other words, I'm no further forward than if I scraped the original web page.
If we want to make the information accessible to a computer, then we need something else. RDF is the obvious way forward.
The difference that RDF makes
To illustrate the difference, let's search for images of the same spider (Enoplognatha latimana) using my Open Search wrapper for Yahoo's images search (described in OpenSearch and iSpecies). Here is the query. This feed is formatted as RSS 1.0, and I can view it in a feed reader, such as NetNewsWire.
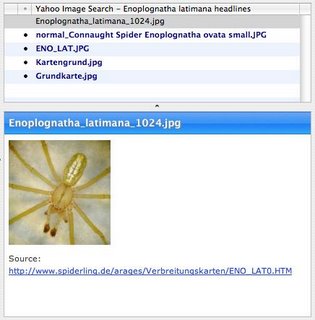
But, because the feed is RSS 1.0 and therefore RDF, the feed contains lots of information on the image in a form that can be easily consumed.
<foaf:Image rdf:about="http://www.spiderling.de/arages/
Fotogalerie/Enoplognatha_latimana_1024.jpg">
<dc:type>image</dc:type>
<dc:title>Enoplognatha_latimana_1024.jpg</dc:title>
<dc:description></dc:description>
<dc:subject>Enoplognatha latimana</dc:subject>
<dc:source>http://www.spiderling.de/arages/
Verbreitungskarten/ENO_LAT0.HTM</dc:source>
<dc:format>image/jpeg</dc:format>
<foaf:thumbnail rdf:resource=
"http://re3.mm-a1.yimg.com/image/206564554"/>
</foaf:Image>
In this example, I use the FOAF and Dublin Core vocabularies. these are widely used, making it easy to integrate this information into a larger database, such as a triple store. To my mind, this is the way forward. We need to move beyond thinking about making data only accessible to people, and making it accessible to computers. Once we do this, then we can start to aggregate and query the huge amounts of data on the web (as exemplified by David's wonderful site on spiders). And once we do that, we may discover all sorts of things that we don't know (see Disconnected databases, and Discovering new things).






 to the Google Scholar results, based on code from
to the Google Scholar results, based on code from  One issue which comes up every so often is how to add data sources to iSpecies. At present iSpecies queries NCBI, Yahoo images, and Google Scholar, each source requiring different code to make the query and handle the response. If adding new sources requires writing code specific to that source then iSpecies would rapidly become a nightmare (leaving aside the issue that until iSpecies is multithreaded, adding additional sources slows everything down -- see
One issue which comes up every so often is how to add data sources to iSpecies. At present iSpecies queries NCBI, Yahoo images, and Google Scholar, each source requiring different code to make the query and handle the response. If adding new sources requires writing code specific to that source then iSpecies would rapidly become a nightmare (leaving aside the issue that until iSpecies is multithreaded, adding additional sources slows everything down -- see  I just noticed this piece written by Jeff Perkel in January who, after "poking around" the iSpecies blog, wrote
I just noticed this piece written by Jeff Perkel in January who, after "poking around" the iSpecies blog, wrote